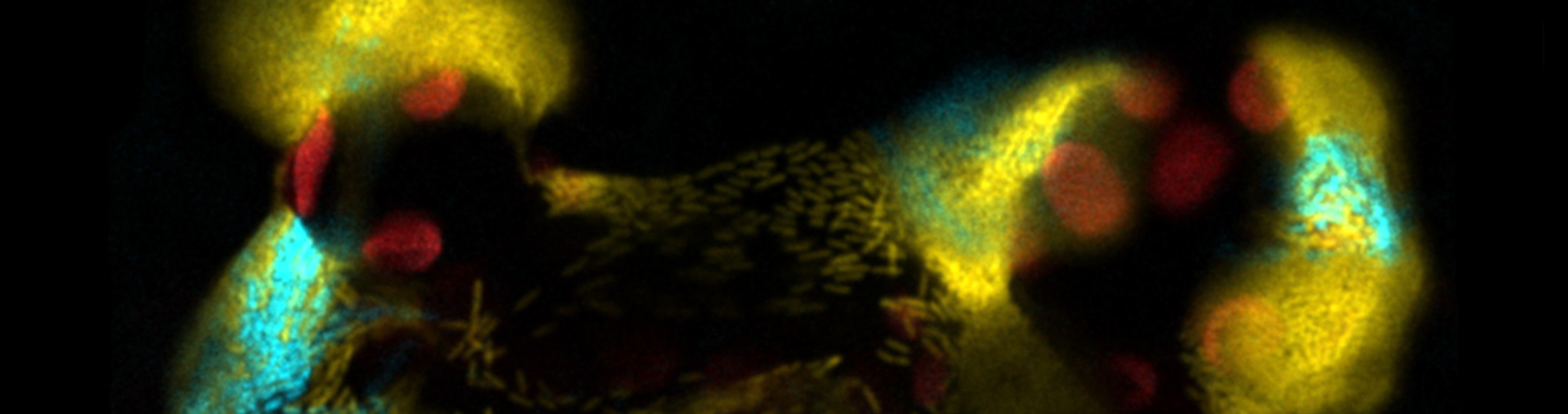
Bacterial pathogens encode virulence systems that facilitate colonization of plant tissue and deliver virulence proteins to suppress plant defenses. Pseudomonas syringae is a phytopathogenic bacterium, relevant both as an academic and agronomic model. The ability of P. syringae to adapt to different niches is an important aspect of its epidemiological success. The virulence of P. syringae relies on its type III secretion system (T3SS) to introduce effectors into the plant host cell, where they can suppress the activation of defense pathways, allowing the pathogen to proliferate and colonize. the leaf apoplast. However, our work has shown that only a part of the bacterial population activates the expression of T3SS genes within the apoplast. Moreover, the expression of T3SS in clonal populations of P. syringae is bistable in a T3SS inducing medium: with a subpopulation of T3SSON bacteria and another of T3SSOFF bacteria. These subpopulations present differences in virulence. Another important asset in the ability of P. syringae to adapt to different niches is the flagellar system, which provides movement and facilitates access to nutrients or to the leaf interior. The flagellar filament is also an inducer of plant defenses. Thus, expression within the apoplast is downregulated. This project will analyze and characterize the expression of the flagellum in the population, inside and outside the plant, and in relation to the expression of T3SS.
Stochastic activation of gene expression in clonal populations can be determined by epigenetic mechanisms, e.g. DNA methylation. We have established the methylomes of two of the main model strains of P. syringae (pathogens of bean and tomato respectively), where we have identified several methylation motifs, different in each strain, abundant in promoter regions and showing an overlap potential binding motifs for transcription factors (TF). The overlap between TF binding sites and methylation is a classic feature of epigenetic regulation of stochastic gene expression. Many of the methylation sites within the genomes of these two P. syringae strains appear hemimethylated or unmethylated, suggesting that they may be involved in gene regulation. In addition, several (partly unmethylated) methylation sites are found within the promoter regions of key T3SS regulators, previously linked by our laboratory to the bistable expression of T3SS genes and likely involved in the regulation between T3SS expression and the flagellar system. This project will characterize the role of DNA methylases in gene regulation in these strains of P. syringae and in the generation of phenotypic heterogeneity of loci related to virulence in this pathogen.
Stochastic activation of gene expression generates phenotypic heterogeneity in the absence of genetic differences and may have adaptive value. We will also investigate the role that different subpopulations play in plant adaptation in different defense contexts and pathosystems, and whether these are important for plant resistance or pathogen resistance to other control strategies.
Code
PID2021-127245OB-I00
DURATION
1 septiembre 2022 – 31 agosto 2025
Funding agency
MCIN/ AEI/10.13039/501100011033/
INVESTIGADOR/A PRINCIPAL
Carmen Beuzón López & Francisco Javier Ruiz Albert
PARTICIPANTES
Javier Ruiz Albert




