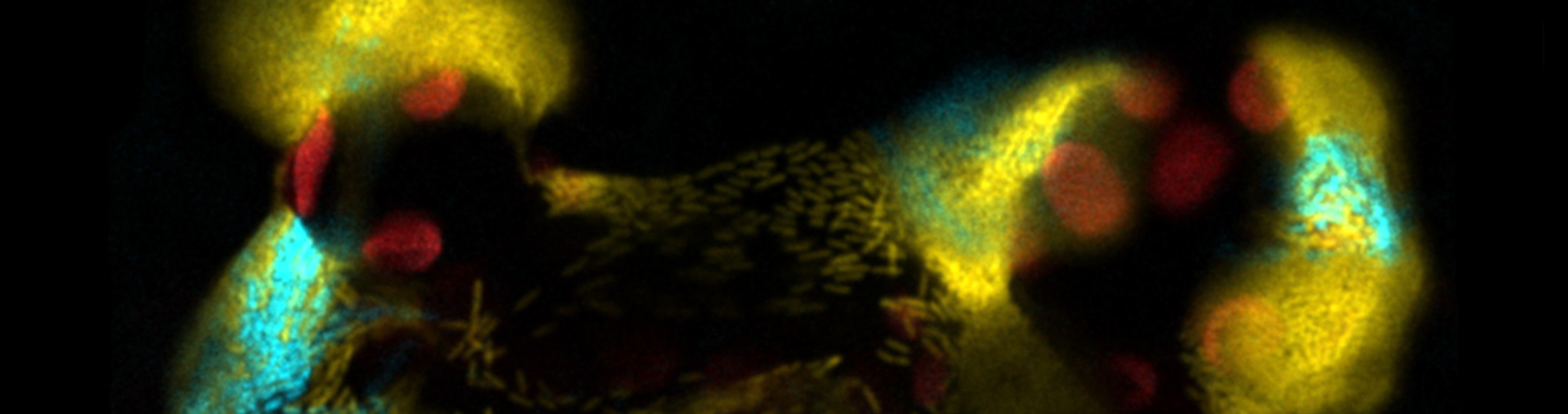
Pseudomonas syringae is very relevant plant pathogenic bacteria, both as a model for the study of plant-pathogen interaction, and for the diseases it causes in a large number of economically important crops. P. syringae reaches the surface of the leaves through rain, subsequently invading the intercellular space (apoplast), where it proliferates suppressing the plant immune system through a set of proteins (effectors) that introduces into the host cell using a type III secretion system (T3SS). Effectors can suppress the two main layers of defense: the first, activated upon recognition of highly conserved molecules known as PAMPs, and the second triggered by specific recognition of pathogen effectors. Activation of either determines the establishment of an immune response in distal leaves known as SAR. Our group was the first to identify the ability to suppress SAR on an effector, HopZ1a. We have shown that HopZ1a interacts with a MAP kinase (ZIP1), involved in the activation of defenses against P. syringae, acetylating key functional residues. Our first objective is the analysis of the molecular mechanisms involved in the activation of defenses by ZIP1 and its suppression by HopZ1a. Our working hypothesis is that ZIP1 regulates the activation of SAR through the regulation of long distance ROS-dependent signaling, following a mechanism similar to their counterparts in animal pathogens The second objective also pursues the analysis of molecular mechanisms, in this case those involved in the formation of bacterial lineages during colonization of the plant. Our team was the first to show that the expression of the T3SS and effectors is bistable during growth in plant. In other words, clonal populations of the pathogen bifurcate into subpopulations or differentiated lineages, expressing or not the T3SS and its secreted effectors. This process is dynamic, reversible and non-genetic, and determines significant differences in virulence. This objective includes the analysis of bacterial and host mechanisms involved in the formation of lineages of P. syringae, which can determine heterogeneity in the activation and suppression of defenses against the pathogen. It also includes the analysis of the impact on the formation of lineages of the treatment with phytosanitary agents against P. syringae, and the importance of this process in the resistance or tolerance to the treatment. Lastly, it also includes analyzing the formation of lineages in the human pathogen Salmonella enterica during colonization of plants. The working hypothesis is that the formation of lineages of P. syringae is important for colonization of the host, and potentially involved in persistence against pathogen control treatments, in a similar manner to what happens in Salmonella and other animal pathogens, and that the plant is a good model for the analysis of the mechanisms involved in the formation of bacterial lineages within the host. The proposed objectives deal with different and very innovative aspects of the plant-pathogen interaction, were first described by our team and are related to the T3SS function and important for the adaptation of the pathogen to the host.
Code
BIO2015-64391-R
Duration
1 enero 2016 – 31 diciembre 2018
Funding agency
MINECO
Principal Investigator
Carmen Beuzón López & Javier Ruiz Albert
Participants
Javier Rueda Blanco




