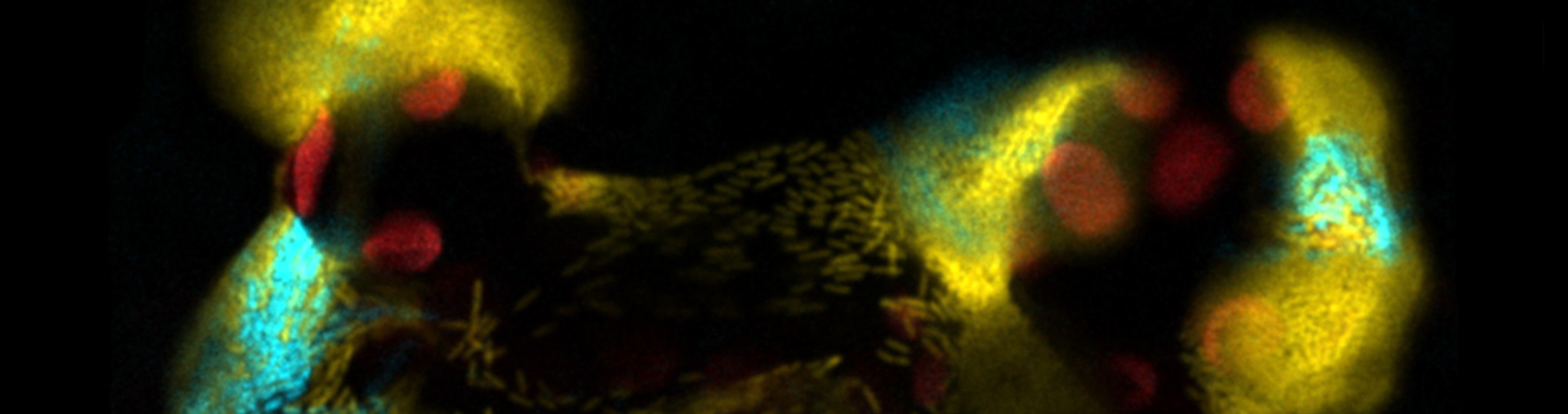
Plants are constantly exposed to potential pathogens. The co-evolution of plant pathogens and their hosts has resulted in highly adapted microbial invasion and colonization strategies, with bacterial systems dedicated to access and colonize the plant, and to deliver virulence proteins to evade the corresponding plant counter-defense mechanisms. This proposal focuses on the Pseudomonas syringae – plant interaction model, relevant both as a basic research and agronomical model. The main virulence assets of P. syringae are the T3SS which delivers virulence proteins (effectors) and the flagellar system that provides movement for access and colonization of intracellular spaces. Yet both systems are targets of the plant counter-defense mechanisms. Therefore, their regulation by the invading bacterial populations is key for the development of disease. We have described for the first time in a plant pathogen the existence of phenotypic heterogeneity in the expression of both the T3SS and the flagellum that leads to the formation of phenotypic bacterial lineages differing in these key aspects of P. syringae virulence, and is integrated into the bacterial regulatory network through an epigenetic mechanism, likely based on DNA methylation. Such phenomenon is bound to have important adaptative consequences for the bacterial-plant interaction and development of disease. In view of all this, we propose the analysis of the mechanisms underlying phenotypic heterogeneity in P. syringae, and its role in plant adaptation. We will perform a genome-wide analysis of the P. syringae methylome in different in planta and in vitro growth conditions, in order to identify phenotypically heterogeneous loci that will be characterized. We intend to identify and characterize the P. syringae DNA methylases associated to such regulation. Additionally, we will further our characterization of the adaptative roles played by phenotypic heterogeneity during plant colonization by P. syringae and we will explore the biotechnological applications of the above. We will also analyse the role of phenotypic heterogeneity during the recently described plant colonization of the human pathogen Salmonella, responsible for numerous epidemic outbreaks linked to internal contamination of crops grown for human consumption. Finally, we propose the isolation and characterization of bacteriophages in agronomical environments, which will be assayed as means to control and detection of P. syringae and Salmonella in commercial crops.
Code
RTI2018-095069-B-I00
Duration
1 enero 2019 – 30 septiembre 2022
Funding agency
Ministerio de Ciencia, Innovación y Universidades
Principal Investigator
Carmen Beuzón López & Javier Ruiz Albert
Participants
Nieve López Pagán, Javier Rueda Blanco,
Ángel del Espino Pérez




